Clustering for outlier removal in tractography
Keywords: clustering, python, diffusion MRI
The human brain is a highly interconnected network with 86 billion brain cells which are called neurons and each one of them have thousands of connections.
Neurons communicate with each other by sending electrical signals that travel down along axons. One way to map the connections between neurons is to track how water molecules move through the brain, a technique called diffusion weighted magnetic resonance imaging (dMRI). Water can flow down along the axon but is trapped inside of it due to faty insulation. By observing the direction of the travel of water, we can estimate the path between brain regions. We can create a map of the pathways in the brain.
Keep in mind that one unit of three dimensional image we get from the MR scanner contains thousands of neurons and axons as well. So, this image in fact only shows that a higher chance of connection between regions instead of the real ones. This image is like predicting the motorways on earth through a stallite image.
Predicting neuronal pathways in brain with dMRI is called tractography. Here is an example of this image:
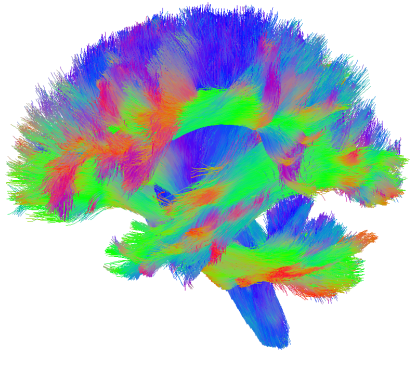
One problem with dMRI is high false positive rates. Connections that do not exist in reality might be estimated with tractography. This is a great talk on this topic Tractography: a story of hope, dreams, …. Inspired from this paper Garyfallidis et al, I created this software for trimming streamlines based on clustering.
This technique is not limited to brain signals only. It can also be used for data that contains trajectoris, for example connections between cities through trainlines or airlines.
In dMRI, we calculate the streamlines that connect brain regions. This streamlines are composed of trajectories in three dimensional space. I used MRtrix to generate whole brain tractography.
First we filter whole brain tractography for specific region of interests (ROIs). Let’s assume we choose region 1 as left motor cortex and region 2 as right motor cortex by using MRtrix function:
connectome2tck tracks_10m_wb.tck assignments_SC.csv edge_1-2.tck -nodes 1,2 -exclusive -files single
Then we load the track files:
tracks = load_tracks(path_track_subj, out_track)
As the streamlines are loaded, we can call the function for clustering and trimming:
streamlines_clean, index_tracks_clean, \
streamlines_outlier, cluster_centroids, cluster_size \
= find_outlier_streamlines(tracks, theta=theta, cutoff_member=cutoff_member)
This function finds outlier streamlines by:
- Resamples all streamlines between two regions so that they will contain equal number of points
- Clusters streamlines based on distance. Here, instead of choosing number of clusters, we choose the maximum distance for the assignment of a streamline to a particular cluster. If the distance is higher than theta, then new cluster is formed. If theta is chosen smaller more streamlines will be discarded.
- Clusters that contain less members than cutoff_member parameter, they are discarded. We chose optimum parameters as cutoff_member = 3 and theta = 20.
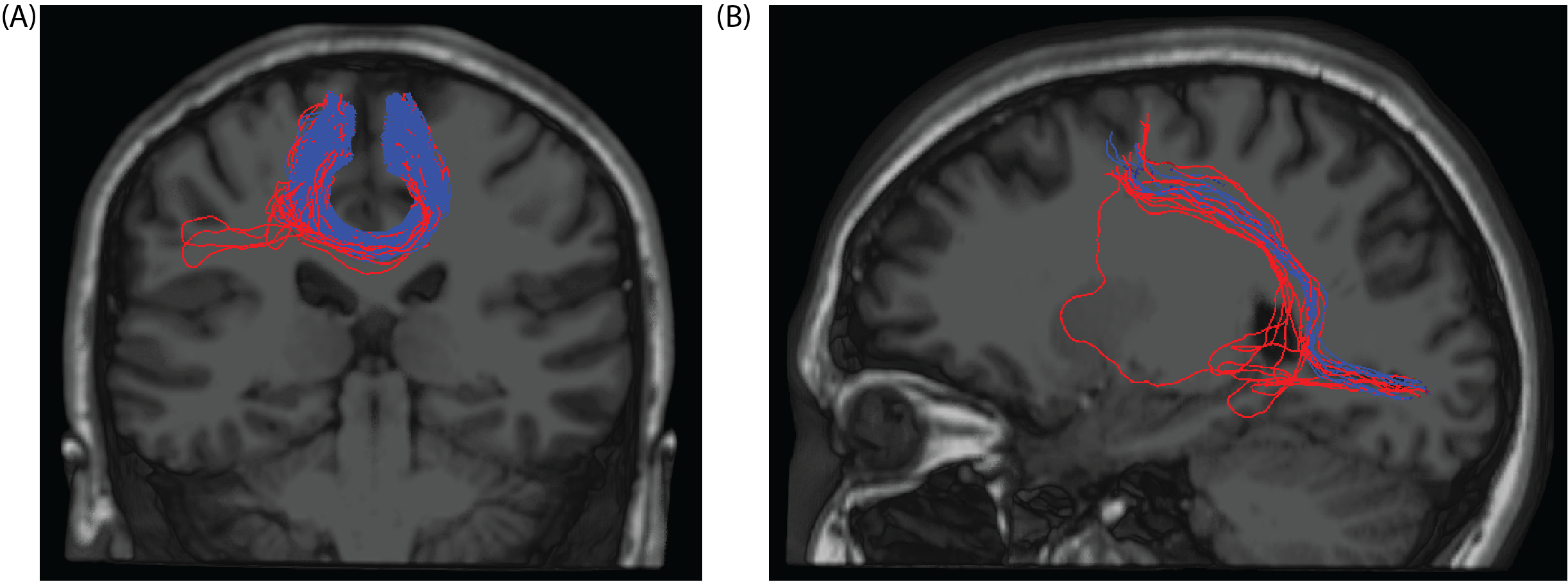
In this figure, we can see two samples for outlier removal. Red lines are the original streamlines. Blue lines are the streamlines that are kept after outlier removal. Since red lines were further than the threshold we chose to cluster centers, they are marked as outliers.
Codes are available at Github.
